J Biomed 2018; 3:13-18. doi:10.7150/jbm.22760 This volume Cite
Review
The Future of Forensic Biology
Division of Biology, State Forensic Science Laboratory, Ministry of Home Affairs, Government of Jharkhand, Near Birsa Munda Jail, Hotwar, Ranchi, Jharkhand, India- 835217.
Received 2017-9-9; Accepted 2017-12-2; Published 2018-1-23
Abstract
The recent technological and scientific development in the field of forensic biology is certainly stamping a huge impact in capturing and convicting the criminals without any doubt. This review enlists forthcoming emerging technologies in forensic biology which would become inevitable and routine tasks in laboratories for solving extremely challenging and critical cases. These include touch DNA profiling, forensic phenotyping, 3D facial reconstruction and morphometrics, microbial forensics, virtual autopsy, DNA methylation and next-generation sequencing methods which would prove boon sooner or later for the crime investigators in finding missing persons, nabbing murderers and sexual assaulters and solving an array of cold cases.
Keywords: Next generation forensic biology, Touch DNA, Forensic phenotyping, Morphometrics, Microbial forensics, Virtopsy, DNA methylation
Introduction
The forensic science today has been expanded with numerous innovative technological advances and molecular discoveries. Following refinement and rigorous testing, many methods/technologies have been adopted by forensic laboratories, including polymerase chain reaction (PCR) [1, 2], capillary electrophoretic instrumentation (Genetic Analyzers) [3, 4], automated liquid handling (Microfluidic devices) [5-7], and expert software systems [8-11]. Introduction of robust validation processes has a long term impact to test evidentiary samples to be presented to the court of law. For example, the credibility of DNA has resulted in DNA becoming the gold standard for forensics and the most effective crime fighting tool available to law enforcement [12].
The progression from Sanger-type sequencing (STS) using dideoxynucleotide triphosphates (ddNTPs) to next generation sequencing (NGS) has further made an impact in the field of forensic science, other than in human health areas such as regenerative or personalized medicine [13, 14]. Today, sequencing of mitochondrial (mtDNA) is already a validated, widely accepted and utilized tool which is helping to resolve many complex cases [15, 16]. Advances in forensic biology, through the introduction of next generation systems in photographical, biological, serological and genetic analyses, are the only ways through which more direct, indirect, corroborative or additive evidences can be collected to be presented in the court of law. The forensic laboratories are expected to migrate in near future from today's current methods to more advanced and efficient technological platforms (Figure 1). In addition, forensic facilities that offer specialized testing for missing persons and mass disaster victim identification will gain great capabilities leveraging these next generation testing systems. Here is the list of advancing technologies that proffer to solve many inapprehensible, recalcitrant and critical cases without any doubt.
Beyond fingerprint Analysis
The fingerprints/footprints primarily consist of organic/inorganic materials [17] or sometimes fats secreted from the skin which could reveal a horde of information about the persons who left them. In many instances, some skin cells could be dislodged on the object while being handled as such or touched with any part of the body. In the future, fingerprints' DNA (touch DNA) from these leftover skin cells on various surfaces can be analyzed through atomic force microscope [18]or even can be collected through optical tweezers equipped with SteREO Lumar.V12 stereomicroscope and UV unit [19], followed by amplification either through AmpFlSTR® NGM™ Amplification Kit (Applied Biosystems) [20] or ploning through φ-29 polymerase [21]. These technologies which helps to fish out touch DNA are therefore potentially promising to identify the traces biological evidence and could be used to develop of genetic profiles from incomprehensible/single cell human samples (theoretically as well as practically). This will allow the crime investigators to identify and correctly nab the culprit in the court of law.
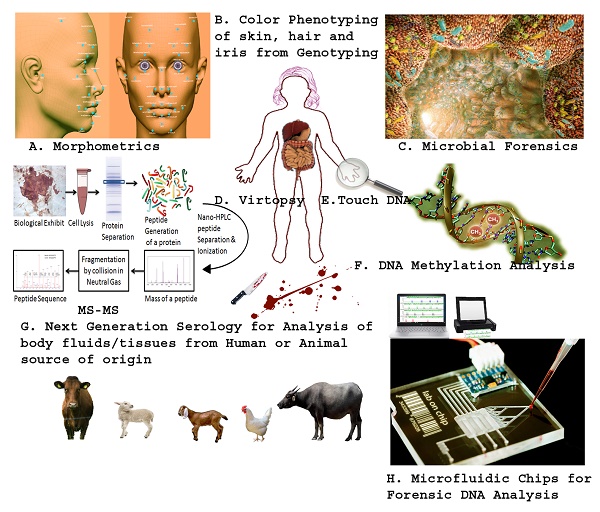
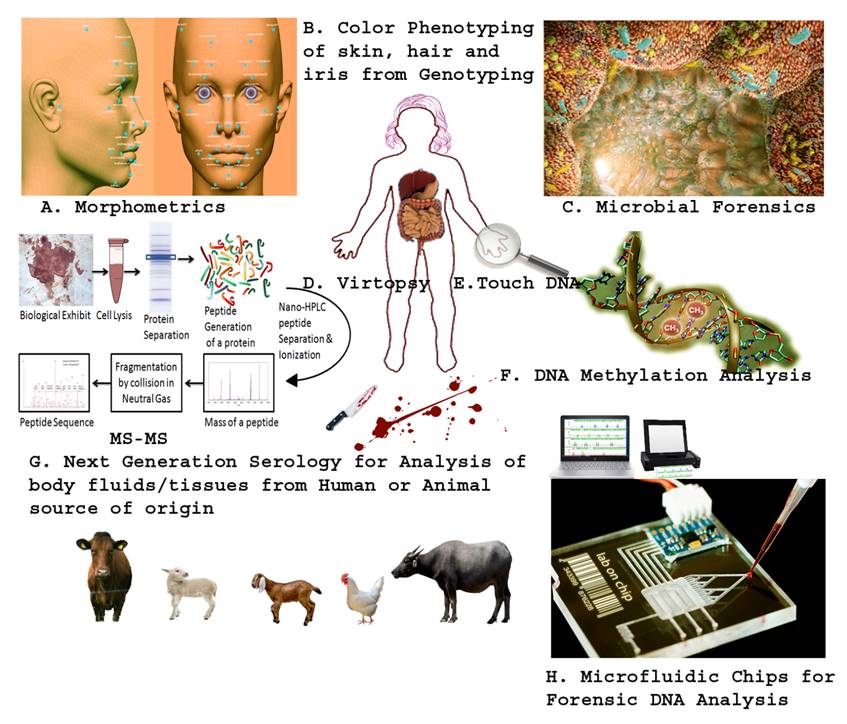
The Next Generation Technologies/Methods in Forensic Biology. A. Morphometric analysis of skull and photographs using next generation 3-D facial reconstruction software using sophisticated laptops. B. Prediction of the color of skin, hair as well iris by sequencing/identifying the genes that actually code for the particular trait/color. C. Study of microbes from human body especially from pubic hair after sexual assault which establishes the exchange of microbial flora in between them. D. Virtual autopsy (Virtopsy®) is a non-invasive method to collect the images of the body through µCT and µMRI to be used in future when may be further required in medico-legal procedures. E. Touch DNA is trace amount of foreign DNA recovered from the person's body during the course of hard press, strangling or kiss from another person. F. DNA methylation analysis is a new method being developed to predict the age of persons, discern monozygotic twins as well as identify various biological fluids recovered from scene of crime. G. Next generation serology involves the use of mass spectrometry especially tandem MS (protein sequencing) which will reveal the origin of the source (Human vs. Animal) through matching the obtained sequence with the non-redundant protein databases, such as UNIPROT. H. Microfluidic chips (for purification, amplification and capillary electrophoresis of DNA) have been developed which are so handy and time efficient that DNA profile of the source of biological exhibit can be established at the scene of crime within a day.
From Genotyping to Forensic Phenotyping
Human phenotype such as hair color, iris color and skin color can be predicted with the type of single nucleotide polymorphism (SNPs) harbored by coding (SLC45A2, SLC24A5, and MC1R)) as well as some non-coding (HERC2 and ASIP) genes in the human genome [22]. Alleles of other pigmentation genes such as TYRP1, DCT and many more have been statistically associated with human iris pigmentation or other human pigmentation. These SNPs data have been translated by a biotech company (Thermo Fisher Scientific) to develop a forensic phenoytyping system called HIrisPlex which employs 24 DNA variants that helps to predict eye and hair color along with six genetic markers which helps to predict blonde hair 69.5 percent of the time, brown hair 78.5 percent of the time, red hair 80 percent of the time, and black hair 87.5 percent of the time [23-25]. This system is based on single base extension of primers using a fluorescently labelled ddNTP terminator ( SNaPshotTM chemistry) in developing polymorphic SNPs profiles of individuals using Genetic Analyzer (Applied Biosystems, Foster City, CA ) [25]. After inclusion of human skin color variant alleles (SNPs), this phenotyping system is palpably going to become an important instrument for routine investigation of questioned human color in the near future of forensics.
Facial Recognition Software and Morphometrics
Automated facial recognition software can be used to identify a victim in disaster or match an accused to a police database [26, 27]. If a photo database of good quality exists such as Interstate Photo System (IPS) possessed by FBI's Next Generation Identification System (NIS) in a utopian police system, then by using a sophisticated laptop one can search a victim/suspect with facial recognition software among millions, barring the stipulation that people face do not change much over time. These softwares are typically used in security systems (biometrics) and can be had from various sources such as
- http://www.bioenabletech.com/tag/free-face-recognition-software/
- https://www.facefirst.com/
- https://facedetection.com/software/
These companies have developed software that recognizes 3D picture of a person's face” using 3D sensors that identifies distinctive features such as globular cranial vault, mandible bone shape, eye sockets and nasal bone. The software's users can also modify a 3D modelled person's attitude or position! For smartphones, the algorithms must be scaled down, which makes them less effective, but in future with more sophistication of mobile phones it may become a handy tool for police officers/forensic scientists to recognize the accused/victim from a crowd [28]. A recent report has analyzed whole genomic sequencing data from different ethnic community around the world (#1061) which is able to identify people according to their physical traits and demography [29]. Morphometrics (the measurement of body shapes) may be used to identify the skeletal remains of missing children, something which is extremely challenging for current forensic experts. Currently, the most commonly used form of morphometric methods are Generalized Procrustes method (GPA), Euclidean Distance Matrix Analysis (EDMA), Principal Components Analysis (PCA) and 3D imaging methods (µMRI and µCT) [30]. One of the major aspect of morphometrics is allometric measurements which compares size of the body parts in relation to the size of the organism [31]. Allometry is also a prominent feature of contour variation for multifarious morphological traits such as craniofacial shape [30].
Microbial Forensics
The most recent report on the ratio of microbiome (largely the bacteria present in our body) to the number of cells in human body is roughly 4:3, of the same order (x1013), and the microbes approximately weighs as 0.2 kg in a typical 70 kg human body [32]. Microbial forensics is a recent discipline in the field of law which employs microbiological methods for analysis of evidence in assorted forms of crimes which includes bioterrorism, hoaxes, accidental release of biological agent/toxin or even rape cases [33-35]. The vast microbial communities (bacteria, yeast and viruses) remain same except after sex. Therefore in future, the pubic hair recovered from sexual assault suspects if do not contain the roots, then the metagenomic analysis of microbes in the hair may become a substitute of evidence [36]. The microbes and hence the microbial DNA differs in males and females and therefore after sex, the microbes in both the male and the female appear to swap, indicating that a sexual act occurred between a particular man and woman. The microbial forensics has been broadened by metagenomic analysis of the microbes from the surroundings also such as from soil [37] in case of bioterroism/murderous crimes, which may also help in collecting and corroborating evidences to correlate with the incident.
Virtual Autopsy (Virtopsy)
Now the human dead body can be preserved virtually for an infinite time which can help a medico-legal doctor/scientist to examine the body by conducting virtual autopsies even in its absence when for religious, personal, or other reasons, spouses or families sometimes do not want their murdered loved one's remains to be autopsied [38]. Invented and trademarked by Prof. Richard Dirnhofer, the virtopsy are noninvasive, damaging neither the body nor forensic evidence [39, 40]. Virtopsy employs post-mortem computed tomography (PMCT), and is best for gas embolism, pneumothorax pulmonary emphysema and the relationship between the internal path of the instrument of aggression and the entry wound. It is quite expensive, but the cost is expected to decrease in coming near future as virtual autopsies will become rampant in forensics especially in medico-legal arena.
DNA methylation
The field of epigenetics has recently made a landmark entry in the field of forensic sciences for the identification and analysis of forensic samples which otherwise contain exactly the same DNA sequences [64]. DNA methylation particularly 5mC (5-methylcytosine) methylation at the CpG sites, the major epigenetic and chemical change in the DNA [41] which without altering the DNA sequence leads to phenotypic variation among the various tissues, monozygotic twins or among humans has provided a new vista to distinguish cell types or tissues [42, 43], monozygotic twins [44-46], age of humans [47-49], human bio-geographic types [44, 50] or even can be used to predict certain criminal behaviors [51-53]. This 15-dalton mark of methylation in the bases of DNA sequence can help to solve an array of critical cases in forensic biology which when stands gloomy with current polymorphic STR technology. With the advances of next-generation sequencing techniques, blooming of DNA methylation datasets and together with standard epigenetic protocols, the prospect of investigating and solving these critical issues in forensic science is highly promising in the court of law.
Next-generation Serology
In very near future, mass spectrometry is going to be a direct method of measuring the source of origin of the biological forensic samples using proteomic analysis (mass spectroscopy, Electrospray Ionization mass spectrometry (ESI-MS and Tandem MS) [54]. It implies that visual examination of origin of species through precipitin test (Paul Uhlenhuth Test) [55] would be more assertively examined by proteomic analysis of characteristic markers present in bodily fluid or tissues (hard or soft) viz. hemoglobin (blood), α-amylase 1 (Saliva), cornulin, cornifin, and/or involucrin (vaginal fluid), semenogelin, prostate-specific antigen, acid phosphatase (semen), Uromodulin (urine), plunc protein (nasal secretions), and immunoglobulins without hemoglobin, stercobilin (faeces). The sequence of protein derived from tandem MS (MS-MS) would help to identify human as well as non-human tissues/secretions that could be easily searched in freely available non-redundant database of protein such as UNIPROT [56].
Conclusive Remarks and Future Prospective
For forensically questioned samples, unswerving tests and time efficient results can be only expected from laboratories when it is immediately processed using standard protocols and advanced sophisticated instruments. Therefore it presumably summons to call the modernized laboratory very close to the crime scene. The so called portable forensic labs which may require handheld electronic sniffers and microfluidic devices in the toxicology or DNA labs respectively, may allow scientists to conduct the experiment at the very crime scene and present instant results to the relevant court of law. The DNA technologies combined with next-generation sequencing methods will certainly save the time and deliver accuracy in analyzing genomic data obtained from tissues [13, 57]. One of the advancement could include the use of Raman spectroscopy to determine whether a suspicious powder is explosive, eliminating the need to use bleach to destroy such substances along with potential evidence [58]. Other advancement could include use of Fourier Transform Infrared (FTIR) spectroscopy to identify drugs such as cocaine in short span of time which could replace several heavy equipments that require various gases, liquids, and solids [59]. The use of handheld 3-D laser scanners [60, 61]and Near-infrared (Near IR) light scanners [62] can also help scientist/police to identify a potential suspect during an investigation. The microfluidic chips on the other hand allow scientists to reduce contamination and build DNA profile at the scene of crime [5-7, 63]. Portable forensics labs could also be equipped with devices capable of transmitting data obtained through the use of next generation facial recognition software and fingerprint scans to government databases for comparison with stored files of such information. The need of the time is to adopt/upgrade the forensic labs with these advanced feasible technologies which could help to nab the perpetrators and bring justice to the innocents by analyzing the obscure samples which otherwise cannot be processed by regular instruments. This will certainly help to reduce the rate of crime in future.
Conflict of Interest
The author (Rana, A.K.) certifies that there is no academic or financial interest accrued to anyone and no other conflicts of interest exists whatsoever.
Acknowledgements
I sincerely thank State Forensic Science Laboratory as well as Department of Police, State Government of Jharkhand, India for financial assistance in the form of monthly salary for its support in bringing out this review at the State Forensic Science Laboratory Jharkhand, Ranchi, India.
References
1. Kasai K, Mukoyama H. [PCR and its application to forensic science]. Tanpakushitsu Kakusan Koso. 1990;35:3150-6
2. Kasai K, Nakamura Y, White R. Amplification of a variable number of tandem repeats (VNTR) locus (pMCT118) by the polymerase chain reaction (PCR) and its application to forensic science. J Forensic Sci. 1990;35:1196-200
3. Sgueglia JB, Geiger S, Davis J. Precision studies using the ABI prism 3100 genetic analyzer for forensic DNA analysis. Anal Bioanal Chem. 2003;376:1247-54
4. Thormann W, Molteni S, Caslavska J, Schmutz A. Clinical and forensic applications of capillary electrophoresis. Electrophoresis. 1994;15:3-12
5. Gibson-Daw G, Albani P, Gassmann M, McCord B. Rapid microfluidic analysis of a Y-STR multiplex for screening of forensic samples. Anal Bioanal Chem. 2017;409:939-47
6. Han JP, Sun J, Wang L, Liu P, Zhuang B, Zhao L, Liu Y, Li CX. The Optimization of Electrophoresis on a Glass Microfluidic Chip and its Application in Forensic Science. J Forensic Sci. 2017
7. Horsman KM, Bienvenue JM, Blasier KR, Landers JP. Forensic DNA analysis on microfluidic devices: a review. J Forensic Sci. 2007;52:784-99
8. Romodanovskii PO, Barinov E, Malyshev AP. [Time of death software product as a readily available tool for the work of a forensic medical expert at the place of occurrence]. Sud Med Ekspert. 2013;56:38-9
9. McDown RJ, Varol C, Carvajal L, Chen L. In-Depth Analysis of Computer Memory Acquisition Software for Forensic Purposes. J Forensic Sci. 2015;61(Suppl 1):S110-6
10. Haned H, Gill P, Lohmueller K, Inman K, Rudin N. Validation of probabilistic genotyping software for use in forensic DNA casework: Definitions and illustrations. Sci Justice. 2015;56:104-8
11. Holland MM, Parson W. GeneMarker(R) HID: A reliable software tool for the analysis of forensic STR data. J Forensic Sci. 2010;56:29-35
12. Lynch M. God's signature: DNA profiling, the new gold standard in forensic science. Endeavour. 2003;27:93-7
13. Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto Calif). 2013;6:287-303
14. Yang Y, Xie B, Yan J. Application of next-generation sequencing technology in forensic science. Genomics Proteomics Bioinformatics. 2014;12:190-7
15. Melton T, Holland C, Holland M. Forensic Mitochondrial DNA Analysis: Current Practice and Future Potential. Forensic Sci Rev. 2012;24:101-22
16. Butler JM, Levin BC. Forensic applications of mitochondrial DNA. Trends Biotechnol. 1998;16:158-62
17. Girod A, Ramotowski R, Weyermann C. Composition of fingermark residue: a qualitative and quantitative review. Forensic Sci Int. 2012;223:10-24
18. Wang C, Stanciu CE, Ehrhardt CJ, Yadavalli VK. Nanoscale characterization of forensically relevant epithelial cells and surface associated extracellular DNA. Forensic Sci Int. 2017;277:252-58
19. Bruck S, Evers H, Heidorn F, Muller U, Kilper R, Verhoff MA. Single cells for forensic DNA analysis-from evidence material to test tube. J Forensic Sci. 2011;56:176-80
20. Soltyszewski I, Szeremeta M, Skawronska M, Niemcunowicz-Janica A, Pepinski W. Typeability of DNA in Touch Traces Deposited on Paper and Optical Data Discs. Adv Clin Exp Med. 2015;24:437-40
21. Giardina E, Pietrangeli I, Martone C, Zampatti S, Marsala P, Gabriele L, Ricci O, Solla G, Asili P, Arcudi G, Spinella A, Novelli G. Whole genome amplification and real-time PCR in forensic casework. BMC Genomics. 2009;10:159
22. Valenzuela RK, Henderson MS, Walsh MH, Garrison NA, Kelch JT, Cohen-Barak O, Erickson DT, John Meaney F, Bruce Walsh J, Cheng KC, Ito S, Wakamatsu K, Frudakis T, Thomas M, Brilliant MH. Predicting phenotype from genotype: normal pigmentation. J Forensic Sci. 2010;55:315-22
23. Ambers AD, Churchill JD, King JL, Stoljarova M, Gill-King H, Assidi M, Abu-Elmagd M, Buhmeida A, Budowle B. More comprehensive forensic genetic marker analyses for accurate human remains identification using massively parallel DNA sequencing. BMC Genomics. 2016;17:750
24. Draus-Barini J, Walsh S, Pospiech E, Kupiec T, Glab H, Branicki W, Kayser M. Bona fide colour: DNA prediction of human eye and hair colour from ancient and contemporary skeletal remains. Investig Genet. 2013;4:3
25. Walsh S, Kayser M. A Practical Guide to the HIrisPlex System: Simultaneous Prediction of Eye and Hair Color from DNA. Methods Mol Biol. 2016;1420:213-31
26. Mazura JC, Juluru K, Chen JJ, Morgan TA, John M, Siegel EL. Facial recognition software success rates for the identification of 3D surface reconstructed facial images: implications for patient privacy and security. J Digit Imaging. 2011;25:347-51
27. Broach J, Yong R, Manuell ME, Nichols C. Use of Facial Recognition Software to Identify Disaster Victims With Facial Injuries. Disaster Med Public Health Prep. 2017:1-5
28. Kramer KM, Hedin DS, Rolkosky DJ. Smartphone based face recognition tool for the blind. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:4538-41
29. Lippert C, Sabatini R, Maher MC, Kang EY, Lee S, Arikan O, Harley A, Bernal A, Garst P, Lavrenko V, Yocum K, Wong T, Zhu M, Yang WY, Chang C, Lu T, Lee CWH, Hicks B, Ramakrishnan S, Tang H, Xie C, Piper J, Brewerton S, Turpaz Y, Telenti A, Roby RK, Och FJ, Venter JC. Identification of individuals by trait prediction using whole-genome sequencing data. Proc Natl Acad Sci U S A. 2017
30. Hallgrimsson B, Percival CJ, Green R, Young NM, Mio W, Marcucio R. Morphometrics, 3D Imaging, and Craniofacial Development. Curr Top Dev Biol. 2015;115:561-97
31. Klingenberg CP. Size, shape, and form: concepts of allometry in geometric morphometrics. Dev Genes Evol. 2016;226:113-37
32. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533
33. Tucker JB, Koblentz GD. The four faces of microbial forensics. Biosecur Bioterror. 2009;7:389-97
34. Schmedes SE, Sajantila A, Budowle B. Expansion of Microbial Forensics. J Clin Microbiol. 2016;54:1964-74
35. Albert J, Wahlberg J, Leitner T, Escanilla D, Uhlen M. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J Virol. 1994;68:5918-24
36. Tridico SR, Murray DC, Addison J, Kirkbride KP, Bunce M. Metagenomic analyses of bacteria on human hairs: a qualitative assessment for applications in forensic science. Investig Genet. 2014;5:16
37. Khodakova AS, Smith RJ, Burgoyne L, Abarno D, Linacre A. Random whole metagenomic sequencing for forensic discrimination of soils. PLoS One. 2014;9:e104996
38. Dirnhofer R, Jackowski C, Vock P, Potter K, Thali MJ. VIRTOPSY: minimally invasive, imaging-guided virtual autopsy. Radiographics. 2006;26:1305-33
39. Plattner T, Thali MJ, Yen K, Sonnenschein M, Stoupis C, Vock P, Zwygart-Brugger K, Kilchor T, Dirnhofer R. Virtopsy-postmortem multislice computed tomography (MSCT) and magnetic resonance imaging (MRI) in a fatal scuba diving incident. J Forensic Sci. 2003;48:1347-55
40. Thali MJ, Yen K, Schweitzer W, Vock P, Boesch C, Ozdoba C, Schroth G, Ith M, Sonnenschein M, Doernhoefer T, Scheurer E, Plattner T, Dirnhofer R. Virtopsy, a new imaging horizon in forensic pathology: virtual autopsy by postmortem multislice computed tomography (MSCT) and magnetic resonance imaging (MRI)-a feasibility study. J Forensic Sci. 2003;48:386-403
41. Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607-17
42. Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044-56
43. Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, Koltsina M, Nilsson TK, Vilo J, Salumets A, Tonisson N. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54
44. Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, Monk D, Hata K, Marques-Bonet T, Wang L, Esteller M. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363-72
45. Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604-9
46. Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman II, Martin NG, Petronis A. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240-5
47. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115
48. Yi SH, Jia YS, Mei K, Yang RZ, Huang DX. Age-related DNA methylation changes for forensic age-prediction. Int J Legal Med. 2015;129:237-44
49. Bekaert B, Kamalandua A, Zapico SC, Van de Voorde W, Decorte R. Improved age determination of blood and teeth samples using a selected set of DNA methylation markers. Epigenetics. 2015;10:922-30
50. Adkins RM, Krushkal J, Tylavsky FA, Thomas F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res A Clin Mol Teratol. 2011;91:728-36
51. Szyf M. DNA methylation, behavior and early life adversity. J Genet Genomics. 2013;40:331-8
52. Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319-23
53. Szyf M. DNA methylation, the early-life social environment and behavioral disorders. J Neurodev Disord. 2011;3:238-49
54. Van Steendam K, De Ceuleneer M, Dhaenens M, Van Hoofstat D, Deforce D. Mass spectrometry-based proteomics as a tool to identify biological matrices in forensic science. Int J Legal Med. 2012;127:287-98
55. Allison AC, Morton JA. Species specificity in the inhibition of antiglobulin sera; a technique for the identification of human and animal bloods. J Clin Pathol. 1953;6:314-9
56. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 2010; 38(D): 142-8.
57. Zhang Y, Jeltsch A. The application of next generation sequencing in DNA methylation analysis. Genes (Basel). 2010;1:85-101
58. Sil S, Umapathy S. Raman spectroscopy explores molecular structural signatures of hidden materials in depth: Universal Multiple Angle Raman Spectroscopy. Sci Rep. 2014;4:5308
59. Marcelo MC, Mariotti KC, Ferrao MF, Ortiz RS. Profiling cocaine by ATR-FTIR. Forensic Sci Int. 2014;246:65-71
60. Polo ME, Felicisimo AM. Analysis of uncertainty and repeatability of a low-cost 3D laser scanner. Sensors (Basel). 2012;12:9046-54
61. Hennessy RJ, McLearie S, Kinsella A, Waddington JL. Facial surface analysis by 3D laser scanning and geometric morphometrics in relation to sexual dimorphism in cerebral-craniofacial morphogenesis and cognitive function. J Anat. 2005;207:283-95
62. Zhang X, Bloch S, Akers W, Achilefu S. Near-infrared molecular probes for in vivo imaging. Curr Protoc Cytom. 2012 Chapter 12: Unit12 27
63. Bruijns B, van Asten A, Tiggelaar R, Gardeniers H. Microfluidic Devices for Forensic DNA Analysis: A Review. Biosensors (Basel). 2016:6
64. Rana A.K, Crime investigation through DNA methylation analysis. methods and applications in forensics. Egypt J Forensic Sci. 2018:8 (7)
Author contact
![]() Corresponding author: Tel.: +91-8292417706; Fax: +91-0651-2562557. Email: ajay1ranacom
Corresponding author: Tel.: +91-8292417706; Fax: +91-0651-2562557. Email: ajay1ranacom